
What is polymerase chain reaction?
If you have ever worked in a molecular biology laboratory you have likely done a polymerase chain reaction (PCR). PCR is an in vitro method in which a small amount of DNA can be copied many many times in a short time period. PCR was invented in the early 1980s by Kary B. Mullis who later shared a Nobel Prize in Chemistry for his work. Since then PCR has become a standard and essential practice in molecular biology and can be used in a multitude of scientific techniques such as molecular cloning or molecular diagnostics.
Steps of PCR
The beauty of PCR is that it can amplify DNA using only a short list of reagents and several heating and cooling steps. PCR relies on heat resistant DNA polymerase from the thermophilic bacterium, Thermos aquaticus (Taq). Taq polymerase is thus a heat resistant enzyme that can withstand changes in temperature. Taq was first identified in the late 1960s during research at hot springs in Yellowstone National Park.
In addition to Taq DNA polymerase, PCR requires free nucleotides (dNTPs), template DNA to amplify from and unique single stranded DNA primers that bind upstream (5’) and downstream (3’) of the DNA region of interest. Primers are crucial for this process as DNA polymerases require an existing strand of DNA to add nucleotides to.
Using these reagents and a series of heating (denaturing) and cooling (annealing) steps Taq polymerase can copy DNA between the primers using the dNTPs.
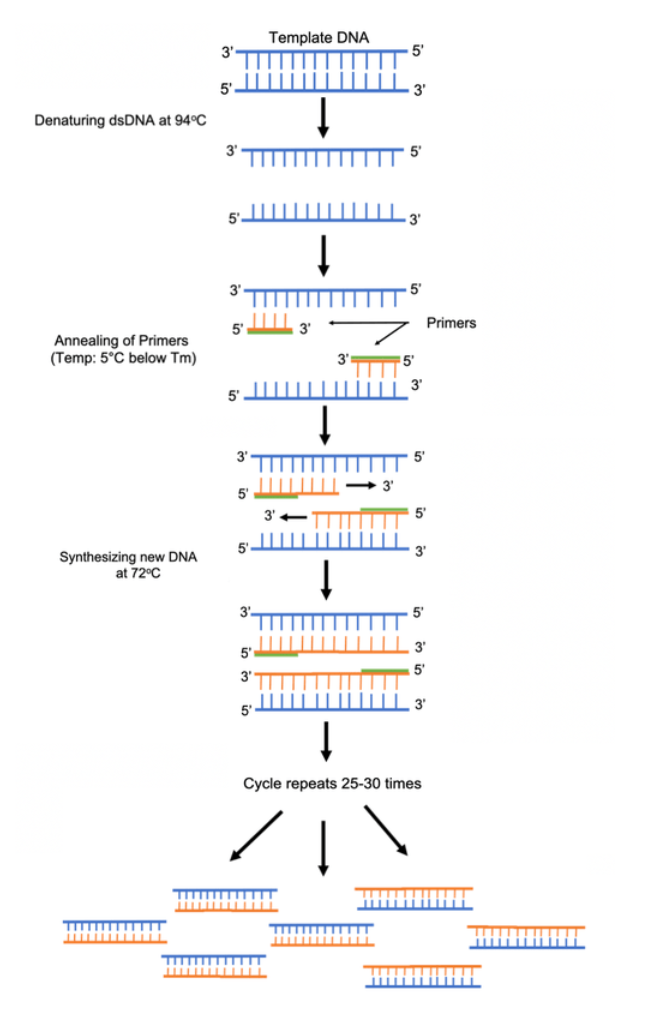 Let’s jump into the specifics of the 3 basic steps of a PCR reaction:
Let’s jump into the specifics of the 3 basic steps of a PCR reaction:
Denaturation- To amplify DNA, the two strands of the template DNA first have to be separated. This occurs by heating the dsDNA template to a point where the hydrogen bonds break between the base pairs. This results in the separation of the two DNA strands.
Annealing- The temperature is then dropped to a range in which the forward and reverse primers are stable. At this temperature the primers can anneal to the single stranded DNA template strands. DNA polymerase is also stable at this temperature and can bind to the primers.
Extension- The temperature is then raised slightly to Taq polymerase’s ideal temperature (70-75oC). At this temperature Taq polymerase can synthesize and elongate the target DNA quickly and accurately.
This process of denaturation, annealing and extension is repeated 25-35 times to exponentially replicate the target DNA of interest. This entire reaction used to be done manually in water baths but is now easily done in a programmed thermocycler. For more details on this procedure check out our PCR protocol page, protocol video and reference on how to design PCR primers.
Types of PCR
Since the invention of PCR, different PCR methods have been developed for different scientific applications. These PCR methods all use the same basic PCR set up and steps but differ in how the PCR products are analyzed.
End point PCR
End point PCR, as the name implies, analyzes the end product of PCR temperature cycling. The final PCR product is often visualized on a diagnostic agarose gel to confirm product presence, size, and relative quantity. End point PCR is most commonly used in molecular cloning, sequencing, and genotyping. It’s extremely useful but is not as quantitative as other methods of PCR. Theoretically scientists should be able to determine the quantity of DNA after a PCR reaction as the amplicon doubles every reaction cycle. However, it is common for dNTPs and other reagents to run low during the final cycles which would slow or stop PCR amplification. Other times, the PCR reaction may not be 100% efficient and produce a reduced amount of product.
End point PCR, which is commonly referred to as simply PCR, is used for an array of different molecular biology techniques and experiments. End point PCR’s goal is to amplify a specific target of DNA. This amplicon can be used in molecular cloning to create a plasmid of interest, for detecting the presence of an insert or piece of DNA in a sample such as in colony PCR, or for generating mutated sequences for site-directed mutagenesis.
